Group Leader: Dr. Sabrina Mühlen
The immune system is designed to defend the body against invaders such as bacteria. Interestingly, while the commensal bacteria that reside in and on our bodies are not recognised as foreign or dangerous, pathogenic bacteria are and hence they induce an immune response. On the other hand, many bacterial pathogens have evolved strategies to escape immune recognition and clearance by actively subverting immune signalling pathways. The innate immune signalling pathways induced by pathogens include pro-inflammatory and cell death signalling pathways as well as autophagy (xenophagy). In our group, we study how pathogenic bacteria subvert the host innate immune response.
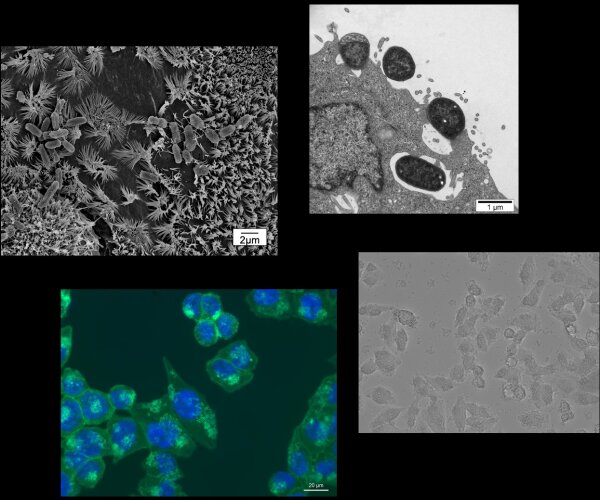
In current research projects, we investigate how the bacterium Staphylococcus aureus induces selective autophagy upon uptake into non-phagocytic cells, then halts autophagy progression and escapes from the autophagosome. We are also aiming to gain a better understanding of how S. aureus adapts to survival inside the host cell as well as of the role autophagy plays during an S. aureus infection. We further study how infection with gastrointestinal pathogens such as pathogenic E. coli and Shigella affects pro-inflammatory signalling, cell death signalling and autophagy and how proteins injected into host cells by these bacteria are involved in the modification of these pathways.
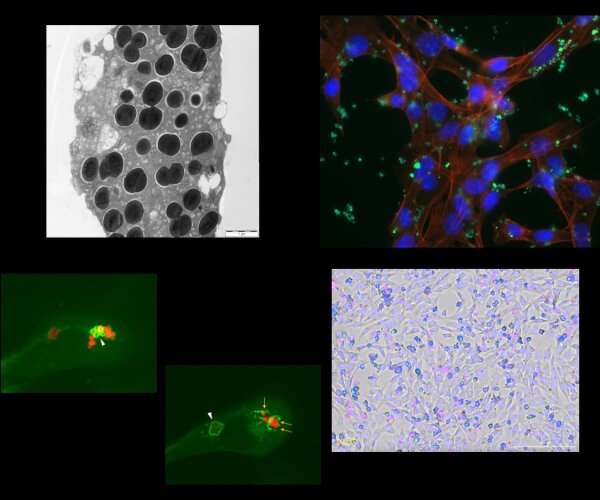
A more thorough understanding of the processes involved in the immune system’s specific response to a certain bacterial pathogen and its clearance as well as insights into the methods employed by the pathogens to interfere with the host’s response will aid the development of future treatment strategies.
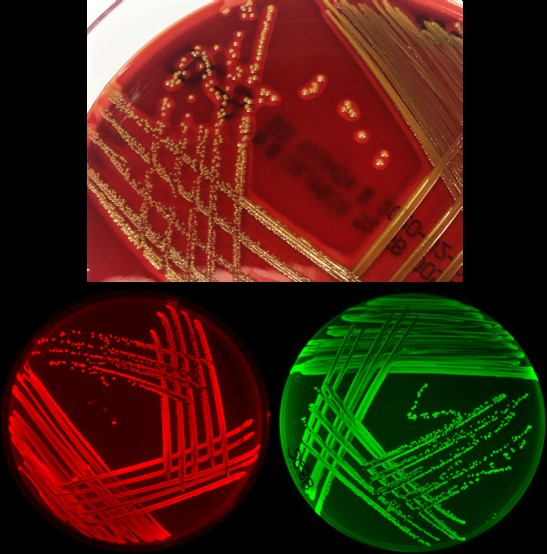

Mühlen S, Heroven AK, Elxnat B, Kahl S, Pieper DH, Dersch P. Infection and antibiotic-associated changes in the fecal microbiota of C. rodentium ϕstx2dact-infected C57BL/6 mice. Antimicrob Agents Chemother. 2024 May 2;68(5):e0005724. doi: 10.1128/aac.00057-24
Blasey N*, Rehrmann D*, Riebisch AK, Mühlen S. Targeting bacterial pathogenesis by inhibiting virulence-associated secretion systems. Review. Front Cell Infect Microbiol. 2023 Jan 10;12:1065561.
Mühlen S, Zapolski'i VA, Bilitewski U, Dersch P. Identification of translocation inhibitors targeting the type III secretion system of enteropathogenic E. coli. Antimicrob Agents Chemother. 2021 Nov 17;65(12):e0095821.
Chaoprasid P*, Lukat P*, Mühlen S*, Heidler T, Gazdag EM, Dong SS, Bi W, Rüter C, Kirchenwitz M, Steffen A, Jänsch L, Stradal TEB, Dersch P* and Blankenfeldt W* (2020). Crystal structure of the bacterial toxin CNFY reveals molecular building blocks for intoxication. EMBO J 2021 Feb 15;40(4):e105202.
Riebisch AK, Mühlen S, Beer YY, Schmitz I (2021). Autophagy – A story of bacteria interfering with the host cell degradation machinery. Invited Review. Pathogens 2021 Jan 22;10(2):110.
Riebisch AK and Mühlen S (2020). Attaching and effacing pathogens: the effector ABC of immune subversion. Invited Review. Future Microbiology. 2020 Jul;15:945-958.
Mühlen S and Dersch P (2020). Treatment strategies for Shiga toxin-producing Escherichia coli infections. Review. Front Cell Infect Microbiol. 2020 May 6;10:169.
Osbelt L, Thiemann S, Smit N, Lesker TR, Schröter M, Galvez EJC, Schmidt-Hohagen K, Pils MC, Mühlen S, Dersch P, Hiller K, Schlüter D, Neumann-Schaal M and Strowig T (2020). Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog 16(3): e1008448.
Mühlen S, Ramming I, Pils MC, Koeppel M, Glaser J, Leong J, Flieger A, Stecher B and Dersch P (2020). Identification of antibiotics that diminish disease in a murine model of enterohemorrhagic Escherichia coli infection. Antimicrob Agents Chemother. 2020 Mar 24;64(4):e02159-19.
Twittenhoff C, Heroven AK, Mühlen S, Dersch P and Narberhaus F (2020). The cnfY RNA thermometer controls the virulence potential of Yersinia pseudotuberculosis. PLoS Pathog 16(1): e1008184.
Lang C, Prager R, Fruth A, Tietze E, Holland G, Laue M, Mühlen S, Dersch P and Flieger A (2018). Widely distributed novel type of pili associated with a Shiga-toxigenic and enteroaggregative E. coli hybrid is essential for aggregative adherence. Emerg Microbes Infect. 2018 Dec 5;7(1):203.
Dersch P, Khan MA, Mühlen S, Görke B. Roles of Regulatory RNAs for Antibiotic Resistance in Bacteria and Their Potential Value as Novel Drug Targets. (2017) Front. Microbiol. 2017 May5;8:803.
Creuzburg K, Giogha C, Wong Fok Lung T, Scott NE, Mühlen S, Hartland EL and Pearson JS (2017). The Type III effector NleD from enteropathogenic E. coli differentiates between host substrates p38 and JNK. Infect Immun. 2017 Jan 26;85(2).
Pearson JS*, Giogha C*, Mühlen S, Nachbur U, Phi CLL, Zhang Y, Hildebrand J, Oates CV, Wong Fok Lung T, Ingle D, Dagley L, Bankovacki A, Petrie E, Schroeder GN, Crepin V, Frankel G, Masters S, Vince J, Murphy J, Sunde M, Webb AI, Silke J, and Hartland EL (2017). A bacterial cysteine protease effector cleaves RHIM proteins to block necroptosis and inflammation. Nat Microbiol. 2017 Jan 13;2:16258.
Zhang Y, Mühlen S, Oates CVL, Pearson JS, Hartland EL (2016). Identification of a distinct substrate binding domain in the bacterial cysteine methyltransferase effectors NleE and OspZ. J Biol Chem. 2016 Sep 16;291(38):20149-62.
Mühlen S and Dersch P (2016). Anti-virulence Strategies to Target Bacterial Infections. Curr Top Microbiol Immunol. 2016;398:147-183.
Giogha C, Wong Fok Lung T, Mühlen S, Pearson JS, Hartland EL (2015). Substrate recognition by the zinc-metalloprotease effector NleC from enteropathogenic Escherichia coli. Cellular Microbiol. Dec;17(12):1766-78.
Nachbur U, Stafford CA, Bankovacki A, Ko H-J, Khakham Y, Fiil BK, Falk H, Zhan Y, Holien JK, Chau D, Hildebrand J, Vince JE, Mühlen S, Kennedy CL, Lowes KN, Murphy J, Gyrd-Hansen M, Parker MW, Hartland EL, Lew AM, Huang DCS, Lessene G and Silke J (2015). A novel RIPK2 inhibitor delays NOD signaling complex formation and transcription factor activation yet prevents inflammatory cytokine production. Nat Commun. 2015 Mar 17;6:6442.
Pearson JS, Giogha C, Ong SY, Kennedy CL, Kelly M, Robinson KS, Wong T, Mansell A, Riedmaier P, Oates CVL, Zaid A, Mühlen S, Crepin VF, Marches O, Ang CS, Williamson NA, O’Reilly LA, Bankovacki A, Nachbur U, Infusini G, Webb A, Silke J, Strasser A, Frankel G, and Hartland EL (2013). A type III effector antagonises death receptor signalling during bacterial gut infection. Nature. 2013 Sep 12;501(7466):247-51.
Ruchaud-Sparagano MH, Mühlen S, and Kenny B (2011). The enteropathogenic E. coli (EPEC) Tir effector protein inhibits NFκB activation by targeting TRAF adaptor proteins. PloS Pathog. 2011 Dec;7(12):e1002414.
Mühlen S, Ruchaud-Sparagano MH, and Kenny B (2010) Proteasome-independent degradation of canonical NFκB complex components by the NleC protein of pathogenic E.coli. J Biol Chem. 2011 Feb 18;286(7):5100-7.
Holmes A, Mühlen S, Roe AJ, Dean P (2010). The EspF effector - a bacterial pathogen's Swiss army knife. Infect Immun. 2010 Nov;78(11):4445-53.
Dean P, Mühlen S, Quitard S, Kenny B (2010). The bacterial effectors EspG and EspG2 induce a destructive calpain activity that is kept in check by the co-delivered Tir effector. Cell Microbiol. 2010 Sep 1;12(9):1308-21.